senic <- read.table('./Datasets/SENIC_data.txt')3 Simple linear regression model: Example
3.1 Read data
3.2 Data pre-processing
colnames(senic) <- c("ID", "LOS", "AGE", "INFRISK", "CULT", "XRAY", "BEDS", "MEDSCHL", "REGION", "CENSUS", "NURSE", "FACS")3.3 Model
Develop a linear regression model to Predict the length of stay based on probability of the person getting infected.
model <- lm(LOS~INFRISK, data = senic)
summary(model)
Call:
lm(formula = LOS ~ INFRISK, data = senic)
Residuals:
Min 1Q Median 3Q Max
-3.0587 -0.7776 -0.1487 0.7159 8.2805
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 6.3368 0.5213 12.156 < 2e-16 ***
INFRISK 0.7604 0.1144 6.645 1.18e-09 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.624 on 111 degrees of freedom
Multiple R-squared: 0.2846, Adjusted R-squared: 0.2781
F-statistic: 44.15 on 1 and 111 DF, p-value: 1.177e-09library(ggplot2)
ggplot(data = senic, aes(x = INFRISK, y = LOS))+
geom_point()+
geom_smooth(method = "lm")`geom_smooth()` using formula = 'y ~ x'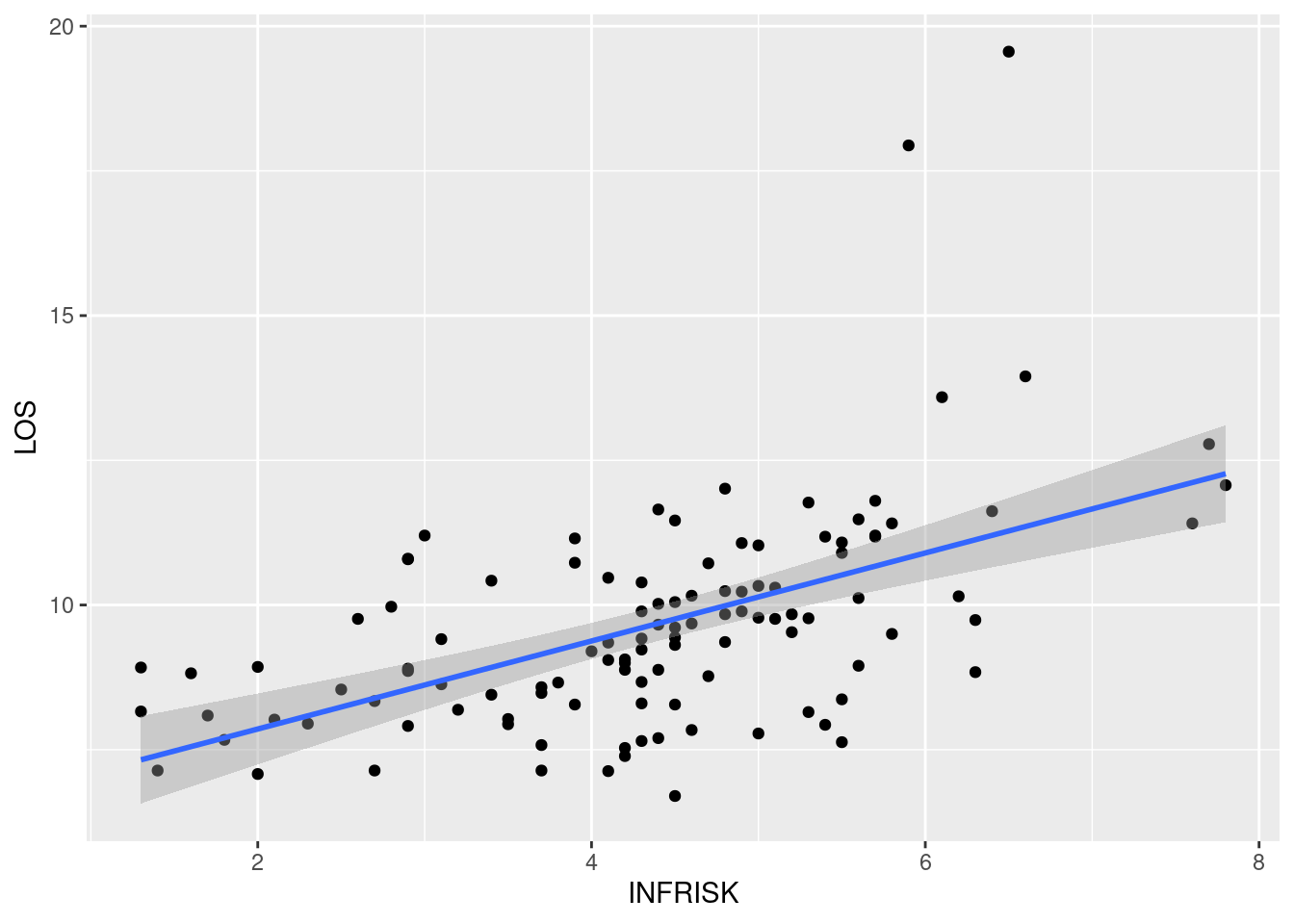
3.4 Error variance
sum(model$residuals**2)/(111)[1] 2.6375183.5 Confidence and Prediction intervals
library(ggplot2)
ci <- predict(model, newdata = senic, interval = "confidence", level = 0.95)
pi <- predict(model, newdata = senic, interval = "prediction", level = 0.95)
data_ci <- cbind(senic, ci)
data_pi <- cbind(senic, pi)
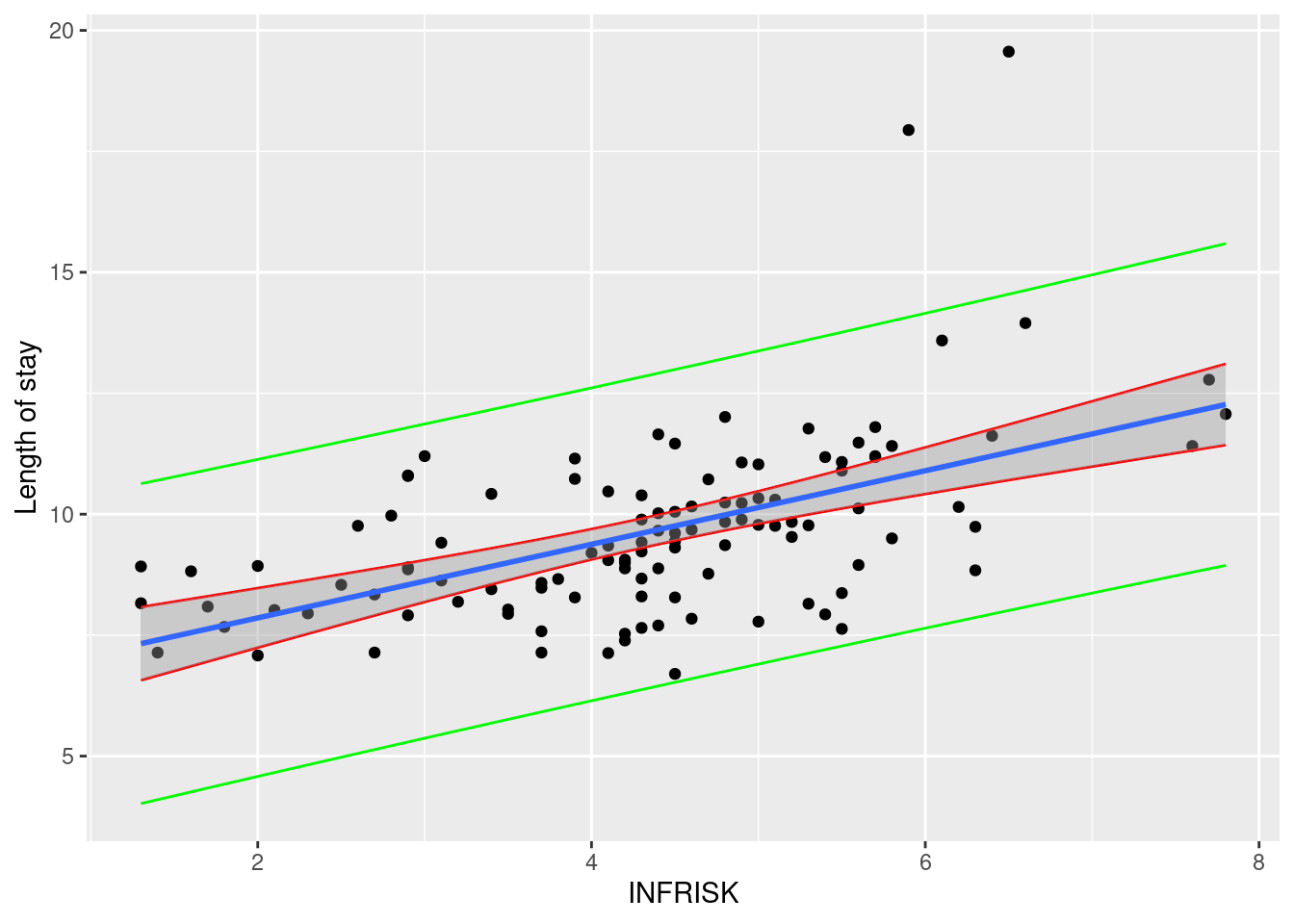
ggplot(data = senic, aes(y = LOS, x = INFRISK))+
geom_point()+
geom_line(data = data_ci, aes(x = INFRISK, y = lwr), color = "red")+
geom_line(data = data_ci, aes(x = INFRISK, y = upr), color = "red")+
geom_line(data = data_pi, aes(x = INFRISK, y = lwr), color = "green")+
geom_line(data = data_pi, aes(x = INFRISK, y = upr), color = "green")+
geom_smooth(method = "lm")+
labs(
y = "Length of stay"
)`geom_smooth()` using formula = 'y ~ x'