import pandas as pd
import numpy as np
import seaborn as sns8 Data wrangling
Data wrangling refers to combining, transforming, and re-arranging data to make it suitable for further analysis. We’ll use Pandas for all data wrangling operations.
8.1 Hierarchical indexing
Until now we have seen only a single level of indexing in the rows and columns of a Pandas DataFrame. Hierarchical indexing refers to having multiple index levels on an axis (row / column) of a Pandas DataFrame. It helps us to work with a higher dimensional data in a lower dimensional form.
8.1.1 Hierarchical indexing in Pandas Series
Let us define Pandas Series as we defined in Chapter 5:
#Defining a Pandas Series
series_example = pd.Series(['these','are','english','words','estas','son','palabras','en','español',
'ce','sont','des','françai','mots'])
series_example0 these
1 are
2 english
3 words
4 estas
5 son
6 palabras
7 en
8 español
9 ce
10 sont
11 des
12 françai
13 mots
dtype: objectLet us use the attribute nlevels to find the number of levels of the row indices of this Series:
series_example.index.nlevels1The Series series_example has only one level of row indices.
Let us introduce another level of row indices while defining the Series:
#Defining a Pandas Series with multiple levels of row indices
series_example = pd.Series(['these','are','english','words','estas','son','palabras','en','español',
'ce','sont','des','françai','mots'],
index=[['English']*4+['Spanish']*5+['French']*5,list(range(1,5))+list(range(1,6))*2])
series_exampleEnglish 1 these
2 are
3 english
4 words
Spanish 1 estas
2 son
3 palabras
4 en
5 español
French 1 ce
2 sont
3 des
4 françai
5 mots
dtype: objectIn the above Series, there are two levels of row indices:
series_example.index.nlevels28.1.2 Hierarchical indexing in Pandas DataFrame
In a Pandas DataFrame, both the rows and the columns can have hierarchical indexing. For example, consider the DataFrame below:
data=np.array([[771517,2697000,815201,3849000],[4.2,5.6,2.8,4.6],
[7.8,234.5,46.9,502],[6749, 597, 52, 305]])
df_example = pd.DataFrame(data,index = [['Demographics']*2+['Geography']*2,
['Population','Unemployment (%)','Area (mile-sq)','Elevation (feet)']],
columns = [['Illinois']*2+['California']*2,['Evanston','Chicago','San Francisco','Los Angeles']])
df_example| Illinois | California | ||||
|---|---|---|---|---|---|
| Evanston | Chicago | San Francisco | Los Angeles | ||
| Demographics | Population | 771517.0 | 2697000.0 | 815201.0 | 3849000.0 |
| Unemployment (%) | 4.2 | 5.6 | 2.8 | 4.6 | |
| Geography | Area (mile-sq) | 7.8 | 234.5 | 46.9 | 502.0 |
| Elevation (feet) | 6749.0 | 597.0 | 52.0 | 305.0 | |
In the above DataFrame, both the rows and columns have 2 levels of indexing. The number of levels of column indices can be found using the attribute nlevels:
df_example.columns.nlevels2The columns attribute will now have a MultiIndex datatype in contrast to the Index datatype with single level of indexing. The same holds for row indices.
type(df_example.columns)pandas.core.indexes.multi.MultiIndexdf_example.columnsMultiIndex([( 'Illinois', 'Evanston'),
( 'Illinois', 'Chicago'),
('California', 'San Francisco'),
('California', 'Los Angeles')],
)The hierarchical levels can have names. Let us assign names to the each level of the row and column labels:
#Naming the row indices levels
df_example.index.names=['Information type', 'Statistic']
#Naming the column indices levels
df_example.columns.names=['State', 'City']
#Viewing the DataFrame
df_example| State | Illinois | California | |||
|---|---|---|---|---|---|
| City | Evanston | Chicago | San Francisco | Los Angeles | |
| Information type | Statistic | ||||
| Demographics | Population | 771517.0 | 2697000.0 | 815201.0 | 3849000.0 |
| Unemployment (%) | 4.2 | 5.6 | 2.8 | 4.6 | |
| Geography | Area (mile-sq) | 7.8 | 234.5 | 46.9 | 502.0 |
| Elevation (feet) | 6749.0 | 597.0 | 52.0 | 305.0 | |
Observe that the names of the row and column labels appear when we view the DataFrame.
8.1.2.1 get_level_values()
The names of the column levels can be obtained using the function get_level_values(). The outer-most level corresponds to the level = 0, and it increases as we go to the inner levels.
#Column levels at level 0 (the outer level)
df_example.columns.get_level_values(0)Index(['Illinois', 'Illinois', 'California', 'California'], dtype='object', name='State')#Column levels at level 1 (the inner level)
df_example.columns.get_level_values(1)Index(['Evanston', 'Chicago', 'San Francisco', 'Los Angeles'], dtype='object', name='City')8.1.3 Subsetting data
We can use the indices at the outer levels to concisely subset a Series / DataFrame.
The first four observations of the Series series_example correspond to the outer row index English, while the last 5 rows correspond to the outer row index Spanish. Let us subset all the observations corresponding to the outer row index English:
#Subsetting data by row-index
series_example['English']1 these
2 are
3 english
4 words
dtype: objectJust like in the case of single level indices, if we wish to subset corresponding to multiple outer-level indices, we put the indices within an additional box bracket []. For example, let us subset all the observations corresponding to the row-indices English and French:
#Subsetting data by multiple row-indices
series_example[['English','French']]English 1 these
2 are
3 english
4 words
French 1 ce
2 sont
3 des
4 françai
5 mots
dtype: objectWe can also subset data using the inner row index. However, we will need to put a : sign to indicate that the row label at the inner level is being used.
#Subsetting data by row-index
series_example[:,2]English are
Spanish son
French sont
dtype: object#Subsetting data by multiple row-indices
series_example.loc[:,[1,2]]English 1 these
Spanish 1 estas
French 1 ce
English 2 are
Spanish 2 son
French 2 sont
dtype: objectAs in Series, we can concisely subset rows / columns in a DataFrame based on the index at the outer levels.
df_example['Illinois']| City | Evanston | Chicago | |
|---|---|---|---|
| Information type | Statistic | ||
| Demographics | Population | 771517.0 | 2697000.0 |
| Unemployment (%) | 4.2 | 5.6 | |
| Geography | Area (mile-sq) | 7.8 | 234.5 |
| Elevation (feet) | 6749.0 | 597.0 |
Note that the dataype of each column name is a tuple. For example, let us find the datatype of the \(1^{st}\) column name:
#First column name
df_example.columns[0]('Illinois', 'Evanston')#Datatype of first column name
type(df_example.columns[0])tupleThus columns at the inner levels can be accessed by specifying the name as a tuple. For example, let us subset the column Evanston:
#Subsetting the column 'Evanston'
df_example[('Illinois','Evanston')]Information type Statistic
Demographics Population 771517.0
Unemployment (%) 4.2
Geography Area (mile-sq) 7.8
Elevation (feet) 6749.0
Name: (Illinois, Evanston), dtype: float64#Subsetting the columns 'Evanston' and 'Chicago' of the outer column level 'Illinois'
df_example.loc[:,('Illinois',['Evanston','Chicago'])]| State | Illinois | ||
|---|---|---|---|
| City | Evanston | Chicago | |
| Information type | Statistic | ||
| Demographics | Population | 771517.0 | 2697000.0 |
| Unemployment (%) | 4.2 | 5.6 | |
| Geography | Area (mile-sq) | 7.8 | 234.5 |
| Elevation (feet) | 6749.0 | 597.0 | |
8.1.4 Practice exercise 1
Read the table consisting of GDP per capita of countries from the webpage: https://en.wikipedia.org/wiki/List_of_countries_by_GDP_(nominal)_per_capita .
To only read the relevant table, read the tables that contain the word ‘Country’.
8.1.4.1
How many levels of indexing are there in the rows and columns?
dfs = pd.read_html('https://en.wikipedia.org/wiki/List_of_countries_by_GDP_(nominal)_per_capita', match = 'Country')
gdp_per_capita = dfs[0]
gdp_per_capita.head()| Country/Territory | UN Region | IMF[4] | World Bank[5] | United Nations[6] | ||||
|---|---|---|---|---|---|---|---|---|
| Country/Territory | UN Region | Estimate | Year | Estimate | Year | Estimate | Year | |
| 0 | Liechtenstein * | Europe | — | — | 169049 | 2019 | 180227 | 2020 |
| 1 | Monaco * | Europe | — | — | 173688 | 2020 | 173696 | 2020 |
| 2 | Luxembourg * | Europe | 127673 | 2022 | 135683 | 2021 | 117182 | 2020 |
| 3 | Bermuda * | Americas | — | — | 110870 | 2021 | 123945 | 2020 |
| 4 | Ireland * | Europe | 102217 | 2022 | 99152 | 2021 | 86251 | 2020 |
Just by looking at the DataFrame, it seems as if there are two levels of indexing for columns and one level of indexing for rows. However, let us confirm it with the nlevels attribute.
gdp_per_capita.columns.nlevels2Yes, there are 2 levels of indexing for columns.
gdp_per_capita.index.nlevels1There is one level of indexing for rows.
8.1.4.2
Subset a DataFrame that selects the country, and the United Nations’ estimates of GDP per capita with the corresponding year.
gdp_per_capita.loc[:,['Country/Territory','United Nations[6]']]| Country/Territory | United Nations[6] | ||
|---|---|---|---|
| Country/Territory | Estimate | Year | |
| 0 | Liechtenstein * | 180227 | 2020 |
| 1 | Monaco * | 173696 | 2020 |
| 2 | Luxembourg * | 117182 | 2020 |
| 3 | Bermuda * | 123945 | 2020 |
| 4 | Ireland * | 86251 | 2020 |
| ... | ... | ... | ... |
| 217 | Madagascar * | 470 | 2020 |
| 218 | Central African Republic * | 481 | 2020 |
| 219 | Sierra Leone * | 475 | 2020 |
| 220 | South Sudan * | 1421 | 2020 |
| 221 | Burundi * | 286 | 2020 |
222 rows × 3 columns
8.1.4.3
Subset a DataFrame that selects only the World Bank and United Nations’ estimates of GDP per capita without the corresponding year or country.
gdp_per_capita.loc[:,(['World Bank[5]','United Nations[6]'],'Estimate')]| World Bank[5] | United Nations[6] | |
|---|---|---|
| Estimate | Estimate | |
| 0 | 169049 | 180227 |
| 1 | 173688 | 173696 |
| 2 | 135683 | 117182 |
| 3 | 110870 | 123945 |
| 4 | 99152 | 86251 |
| ... | ... | ... |
| 217 | 515 | 470 |
| 218 | 512 | 481 |
| 219 | 516 | 475 |
| 220 | 1120 | 1421 |
| 221 | 237 | 286 |
222 rows × 2 columns
8.1.4.4
Subset a DataFrame that selects the country and only the World Bank and United Nations’ estimates of GDP per capita without the corresponding year or country.
gdp_per_capita.loc[:,[('Country/Territory','Country/Territory'),('United Nations[6]','Estimate'),('World Bank[5]','Estimate')]]| Country/Territory | United Nations[6] | World Bank[5] | |
|---|---|---|---|
| Country/Territory | Estimate | Estimate | |
| 0 | Liechtenstein * | 180227 | 169049 |
| 1 | Monaco * | 173696 | 173688 |
| 2 | Luxembourg * | 117182 | 135683 |
| 3 | Bermuda * | 123945 | 110870 |
| 4 | Ireland * | 86251 | 99152 |
| ... | ... | ... | ... |
| 217 | Madagascar * | 470 | 515 |
| 218 | Central African Republic * | 481 | 512 |
| 219 | Sierra Leone * | 475 | 516 |
| 220 | South Sudan * | 1421 | 1120 |
| 221 | Burundi * | 286 | 237 |
222 rows × 3 columns
8.1.4.5
Drop all columns consisting of years. Use the level argument of the drop() method.
gdp_per_capita = gdp_per_capita.drop(columns='Year',level=1)
gdp_per_capita| Country/Territory | UN Region | IMF[4] | World Bank[5] | United Nations[6] | |
|---|---|---|---|---|---|
| Country/Territory | UN Region | Estimate | Estimate | Estimate | |
| 0 | Liechtenstein * | Europe | — | 169049 | 180227 |
| 1 | Monaco * | Europe | — | 173688 | 173696 |
| 2 | Luxembourg * | Europe | 127673 | 135683 | 117182 |
| 3 | Bermuda * | Americas | — | 110870 | 123945 |
| 4 | Ireland * | Europe | 102217 | 99152 | 86251 |
| ... | ... | ... | ... | ... | ... |
| 217 | Madagascar * | Africa | 522 | 515 | 470 |
| 218 | Central African Republic * | Africa | 496 | 512 | 481 |
| 219 | Sierra Leone * | Africa | 494 | 516 | 475 |
| 220 | South Sudan * | Africa | 328 | 1120 | 1421 |
| 221 | Burundi * | Africa | 293 | 237 | 286 |
222 rows × 5 columns
8.1.4.6
In the dataset obtained above, drop the inner level of the column labels. Use the droplevel() method.
gdp_per_capita = gdp_per_capita.droplevel(1,axis=1)
gdp_per_capita| Country/Territory | UN Region | IMF[4] | World Bank[5] | United Nations[6] | |
|---|---|---|---|---|---|
| 0 | Liechtenstein * | Europe | — | 169049 | 180227 |
| 1 | Monaco * | Europe | — | 173688 | 173696 |
| 2 | Luxembourg * | Europe | 127673 | 135683 | 117182 |
| 3 | Bermuda * | Americas | — | 110870 | 123945 |
| 4 | Ireland * | Europe | 102217 | 99152 | 86251 |
| ... | ... | ... | ... | ... | ... |
| 217 | Madagascar * | Africa | 522 | 515 | 470 |
| 218 | Central African Republic * | Africa | 496 | 512 | 481 |
| 219 | Sierra Leone * | Africa | 494 | 516 | 475 |
| 220 | South Sudan * | Africa | 328 | 1120 | 1421 |
| 221 | Burundi * | Africa | 293 | 237 | 286 |
222 rows × 5 columns
8.1.5 Practice exercise 2
Recall problem 2(e) from assignment 3 on Pandas, where we needed to find the African country that is the closest to country \(G\) (Luxembourg) with regard to social indicators.
We will solve the question with the regular way in which we use single level of indexing (as you probably did during this assignment), and see if it is easier to do with hierarchical indexing.
Execute the code below that we used to pre-process data to make it suitable for answering this question.
#Pre-processing data - execute this code
social_indicator = pd.read_csv("./Datasets/social_indicator.txt",sep="\t",index_col = 0)
social_indicator.geographic_location = social_indicator.geographic_location.apply(lambda x: 'Asia' if 'Asia' in x else 'Europe' if 'Europe' in x else 'Africa' if 'Africa' in x else x)
social_indicator.rename(columns={'geographic_location':'continent'},inplace=True)
social_indicator = social_indicator.sort_index(axis=1)
social_indicator.drop(columns=['region','contraception'],inplace=True)Below is the code to find the African country that is the closest to country \(G\) (Luxembourg) using single level of indexing. Your code in the assignment is probably similar to the one below:
#Finding the index of the country G (Luxembourg) that has the maximum GDP per capita
country_max_gdp_position = social_indicator.gdpPerCapita.argmax()
#Scaling the social indicator dataset
social_indicator_scaled = social_indicator.iloc[:,2:].apply(lambda x: (x-x.mean())/(x.std()))
#Computing the Manhattan distances of all countries from country G (Luxembourg)
manhattan_distances = (social_indicator_scaled-social_indicator_scaled.iloc[country_max_gdp_position,:]).abs().sum(axis=1)
#Finding the indices of African countries
african_countries_indices = social_indicator.loc[social_indicator.continent=='Africa',:].index
#Filtering the Manhattan distances of African countries from country G (Luxembourg)
manhattan_distances_African = manhattan_distances[african_countries_indices]
#Finding the country among African countries that has the least Manhattan distance to country G (Luxembourg)
social_indicator.loc[manhattan_distances_African.idxmin(),'country']'Reunion'8.1.5.1
Use the method set_index() to set continent and country as hierarchical indices of rows. Find the African country that is the closest to country \(G\) (Luxembourg) using this hierarchically indexed data. How many lines will be eliminated from the code above? Which lines will be eliminated?
Hint: Since continent and country are row indices, you don’t need to explicitly find:
The row index of country \(G\) (Luxembourg),
The row indices of African countries.
The Manhattan distances for African countries.
social_indicator.set_index(['continent','country'],inplace = True)
social_indicator_scaled = social_indicator.apply(lambda x: (x-x.mean())/(x.std()))
manhattan_distances = (social_indicator_scaled-social_indicator_scaled.loc[('Europe','Luxembourg'),:]).abs().sum(axis=1)
manhattan_distances['Africa'].idxmin()'Reunion'As we have converted the columns continent and country to row indices, all the lines of code where we were keeping track of the index of country \(G\), African countries, and Manhattan distances of African countries are eliminated. Three lines of code are eliminated.
Hierarchical indexing relieves us from keeping track of indices, if we set indices that are relatable to our analysis.
8.1.5.2
Use the Pandas DataFrame method mean() with the level argument to find the mean value of all social indicators for each continent.
social_indicator.mean(level=0)| economicActivityFemale | economicActivityMale | gdpPerCapita | illiteracyFemale | illiteracyMale | infantMortality | lifeFemale | lifeMale | totalfertilityrate | |
|---|---|---|---|---|---|---|---|---|---|
| continent | |||||||||
| Asia | 41.592683 | 79.282927 | 27796.390244 | 23.635951 | 13.780878 | 39.853659 | 70.724390 | 66.575610 | 3.482927 |
| Africa | 46.732258 | 79.445161 | 7127.483871 | 52.907226 | 33.673548 | 77.967742 | 56.841935 | 53.367742 | 4.889677 |
| Oceania | 51.280000 | 77.953333 | 14525.666667 | 9.666667 | 6.585200 | 23.666667 | 72.406667 | 67.813333 | 3.509333 |
| North America | 45.238095 | 77.166667 | 18609.047619 | 17.390286 | 14.609905 | 22.904762 | 75.457143 | 70.161905 | 2.804286 |
| South America | 42.008333 | 75.575000 | 15925.916667 | 9.991667 | 6.750000 | 34.750000 | 72.691667 | 66.975000 | 2.872500 |
| Europe | 52.060000 | 70.291429 | 45438.200000 | 2.308343 | 1.413543 | 10.571429 | 77.757143 | 70.374286 | 1.581714 |
8.1.6 Practice exercise 3
Let us try to find the areas where NU students lack in diversity. Read survey_data_clean.csv. Use hierarchical indexing to classify the columns as follows:
Classify the following variables as lifestyle:
lifestyle = ['fav_alcohol', 'parties_per_month', 'smoke', 'weed','streaming_platforms', 'minutes_ex_per_week',
'sleep_hours_per_day', 'internet_hours_per_day', 'procrastinator', 'num_clubs','student_athlete','social_media']Classify the following variables as personality:
personality = ['introvert_extrovert', 'left_right_brained', 'personality_type',
'num_insta_followers', 'fav_sport','learning_style','dominant_hand']Classify the following variables as opinion:
opinion = ['love_first_sight', 'expected_marriage_age', 'expected_starting_salary', 'how_happy',
'fav_number', 'fav_letter', 'fav_season', 'political_affliation', 'cant_change_math_ability',
'can_change_math_ability', 'math_is_genetic', 'much_effort_is_lack_of_talent']Classify the following variables as academic information:
academic_info = ['major', 'num_majors_minors',
'high_school_GPA', 'NU_GPA', 'school_year','AP_stats', 'used_python_before']Classify the following variables as demographics:
demographics = [ 'only_child','birth_month',
'living_location_on_campus', 'age', 'height', 'height_father',
'height_mother', 'childhood_in_US', 'gender', 'region_of_residence']Write a function that finds the number of variables having outliers in a dataset. Apply the function to each of the 5 categories of variables in the dataset. Our hypothesis is that the category that has the maximum number of variables with outliers has the least amount of diversity. For continuous variables, use Tukey’s fences criterion to identify outliers. For categorical variables, consider levels having less than 1% observations as outliers. Assume that numeric variables that have more than 2 distinct values are continuous.
Solution:
#Using hierarchical indexing to classify columns
#Reading data
survey_data = pd.read_csv('./Datasets/survey_data_clean.csv')
#Arranging columns in the order of categories
survey_data_HI = survey_data[lifestyle+personality+opinion+academic_info+demographics]
#Creating hierarchical indexing to classify columns
survey_data_HI.columns=[['lifestyle']*len(lifestyle)+['personality']*len(personality)+['opinion']*len(opinion)+\
['academic_info']*len(academic_info)+['demographics']*len(demographics),lifestyle+\
personality+opinion+academic_info+demographics]#Function to identify outliers based on Tukey's fences for continous variables and 1% criterion for categorical variables
def rem_outliers(x):
if ((len(x.value_counts())>2) & (x.dtype!='O')):#continuous variable
q1 =x.quantile(0.25)
q3 = x.quantile(0.75)
intQ_range = q3-q1
#Tukey's fences
Lower_fence = q1 - 1.5*intQ_range
Upper_fence = q3 + 1.5*intQ_range
num_outliers = ((x<Lower_fence) | (x>Upper_fence)).sum()
if num_outliers>0:
return True
return False
else: #categorical variable
if np.min(x.value_counts()/len(x))<0.01:
return True
return False#Number of variables containing outlier(s) in each category
for category in survey_data_HI.columns.get_level_values(0).unique():
print("Number of missing values for category ",category," = ",survey_data_HI[category].apply(rem_outliers).sum())Number of missing values for category lifestyle = 7
Number of missing values for category personality = 2
Number of missing values for category opinion = 4
Number of missing values for category academic_info = 3
Number of missing values for category demographics = 4The lifestyle category has the highest number of variables containing outlier(s). If the hypothesis is true, then NU students have the least diversity in their lifestyle, among all the categories.
Although one may say that the lifestyle category has the the highest number of columns (as shown below), the proportion of columns having outlier(s) is also the highest for this category.
for category in survey_data_HI.columns.get_level_values(0).unique():
print("Number of columns in category ",category," = ",survey_data_HI[category].shape[1])Number of columns in category lifestyle = 12
Number of columns in category personality = 7
Number of columns in category opinion = 12
Number of columns in category academic_info = 7
Number of columns in category demographics = 108.1.7 Reshaping data
Apart from ease in subsetting data, hierarchical indexing also plays a role in reshaping data.
8.1.7.1 unstack() (Pandas Series method)
The Pandas Series method unstack() pivots the desired level of row indices to columns, thereby creating a DataFrame. By default, the inner-most level of the row labels is pivoted.
#Pivoting the inner-most Series row index to column labels
series_example_unstack = series_example.unstack()
series_example_unstack| 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|
| English | these | are | english | words | NaN |
| French | ce | sont | des | françai | mots |
| Spanish | estas | son | palabras | en | español |
We can pivot the outer level of the row labels by specifying it in the level argument:
#Pivoting the outer row indices to column labels
series_example_unstack = series_example.unstack(level=0)
series_example_unstack| English | French | Spanish | |
|---|---|---|---|
| 1 | these | ce | estas |
| 2 | are | sont | son |
| 3 | english | des | palabras |
| 4 | words | françai | en |
| 5 | NaN | mots | español |
8.1.7.2 unstack() (Pandas DataFrame method)
The Pandas DataFrame method unstack() pivots the specified level of row indices to the new inner-most level of column labels. By default, the inner-most level of the row labels is pivoted.
#Pivoting the inner level of row labels to the inner-most level of column labels
df_example.unstack()| State | Illinois | California | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| City | Evanston | Chicago | San Francisco | Los Angeles | ||||||||||||
| Statistic | Area (mile-sq) | Elevation (feet) | Population | Unemployement (%) | Area (mile-sq) | Elevation (feet) | Population | Unemployement (%) | Area (mile-sq) | Elevation (feet) | Population | Unemployement (%) | Area (mile-sq) | Elevation (feet) | Population | Unemployement (%) |
| Information type | ||||||||||||||||
| Demographics | NaN | NaN | 771517.0 | 4.2 | NaN | NaN | 2697000.0 | 5.6 | NaN | NaN | 815201.0 | 2.8 | NaN | NaN | 3849000.0 | 4.6 |
| Geography | 7.8 | 6749.0 | NaN | NaN | 234.5 | 597.0 | NaN | NaN | 46.9 | 52.0 | NaN | NaN | 502.0 | 305.0 | NaN | NaN |
As with Series, we can pivot the outer level of the row labels by specifying it in the level argument:
#Pivoting the outer level (level = 0) of row labels to the inner-most level of column labels
df_example.unstack(level=0)| State | Illinois | California | ||||||
|---|---|---|---|---|---|---|---|---|
| City | Evanston | Chicago | San Francisco | Los Angeles | ||||
| Information type | Demographics | Geography | Demographics | Geography | Demographics | Geography | Demographics | Geography |
| Statistic | ||||||||
| Area (mile-sq) | NaN | 7.8 | NaN | 234.5 | NaN | 46.9 | NaN | 502.0 |
| Elevation (feet) | NaN | 6749.0 | NaN | 597.0 | NaN | 52.0 | NaN | 305.0 |
| Population | 771517.0 | NaN | 2697000.0 | NaN | 815201.0 | NaN | 3849000.0 | NaN |
| Unemployement (%) | 4.2 | NaN | 5.6 | NaN | 2.8 | NaN | 4.6 | NaN |
8.1.7.3 stack()
The inverse of unstack() is the stack() method, which creates the inner-most level of row indices by pivoting the column labels of the prescribed level.
Note that if the column labels have only one level, we don’t need to specify a level.
#Stacking the columns of a DataFrame
series_example_unstack.stack()English 1 these
2 are
3 english
4 words
French 1 ce
2 sont
3 des
4 françai
5 mots
Spanish 1 estas
2 son
3 palabras
4 en
5 español
dtype: objectHowever, if the columns have multiple levels, we can specify the level to stack as the inner-most row level. By default, the inner-most column level is stacked.
#Stacking the inner-most column labels inner-most row indices
df_example.stack()| State | California | Illinois | ||
|---|---|---|---|---|
| Information type | Statistic | City | ||
| Demographics | Population | Chicago | NaN | 2697000.0 |
| Evanston | NaN | 771517.0 | ||
| Los Angeles | 3849000.0 | NaN | ||
| San Francisco | 815201.0 | NaN | ||
| Unemployement (%) | Chicago | NaN | 5.6 | |
| Evanston | NaN | 4.2 | ||
| Los Angeles | 4.6 | NaN | ||
| San Francisco | 2.8 | NaN | ||
| Geography | Area (mile-sq) | Chicago | NaN | 234.5 |
| Evanston | NaN | 7.8 | ||
| Los Angeles | 502.0 | NaN | ||
| San Francisco | 46.9 | NaN | ||
| Elevation (feet) | Chicago | NaN | 597.0 | |
| Evanston | NaN | 6749.0 | ||
| Los Angeles | 305.0 | NaN | ||
| San Francisco | 52.0 | NaN |
#Stacking the outer column labels inner-most row indices
df_example.stack(level=0)| City | Chicago | Evanston | Los Angeles | San Francisco | ||
|---|---|---|---|---|---|---|
| Information type | Statistic | State | ||||
| Demographics | Population | California | NaN | NaN | 3849000.0 | 815201.0 |
| Illinois | 2697000.0 | 771517.0 | NaN | NaN | ||
| Unemployement (%) | California | NaN | NaN | 4.6 | 2.8 | |
| Illinois | 5.6 | 4.2 | NaN | NaN | ||
| Geography | Area (mile-sq) | California | NaN | NaN | 502.0 | 46.9 |
| Illinois | 234.5 | 7.8 | NaN | NaN | ||
| Elevation (feet) | California | NaN | NaN | 305.0 | 52.0 | |
| Illinois | 597.0 | 6749.0 | NaN | NaN |
8.2 Merging data
The Pandas DataFrame method merge() uses columns defined as key column(s) to merge two datasets. In case the key column(s) are not defined, the overlapping column(s) are considered as the key columns.
8.2.1 Join types
When a dataset is merged with another based on key column(s), one of the following four types of join will occur depending on the repetition of the values of the key(s) in the datasets.
- One-to-one, (ii) Many-to-one, (iii) One-to-Many, and (iv) Many-to-many
The type of join may sometimes determine the number of rows to be obtained in the merged dataset. If we don’t get the expected number of rows in the merged dataset, an investigation of the datsets may be neccessary to identify and resolve the issue. There may be several possible issues, for example, the dataset may not be arranged in a way that we have assumed it to be arranged.
We’ll use toy datasets to understand the above types of joins. The .csv files with the prefix student consist of the names of a few students along with their majors, and the files with the prefix skills consist of the names of majors along with the skills imparted by the respective majors.
data_student = pd.read_csv('./Datasets/student_one.csv')
data_skill = pd.read_csv('./Datasets/skills_one.csv')8.2.1.1 One-to-one join
Each row in one dataset is linked (or related) to a single row in another dataset based on the key column(s).
data_student| Student | Major | |
|---|---|---|
| 0 | Kitana | Statistics |
| 1 | Jax | Computer Science |
| 2 | Sonya | Material Science |
| 3 | Johnny | Music |
data_skill| Major | Skills | |
|---|---|---|
| 0 | Statistics | Inference |
| 1 | Computer Science | Machine learning |
| 2 | Material Science | Structure prediction |
| 3 | Music | Opera |
pd.merge(data_student,data_skill)| Student | Major | Skills | |
|---|---|---|---|
| 0 | Kitana | Statistics | Inference |
| 1 | Jax | Computer Science | Machine learning |
| 2 | Sonya | Material Science | Structure prediction |
| 3 | Johnny | Music | Opera |
8.2.1.2 Many-to-one join
One or more rows in one dataset is linked (or related) to a single row in another dataset based on the key column(s).
data_student = pd.read_csv('./Datasets/student_many.csv')
data_skill = pd.read_csv('./Datasets/skills_one.csv')data_student| Student | Major | |
|---|---|---|
| 0 | Kitana | Statistics |
| 1 | Kitana | Computer Science |
| 2 | Jax | Computer Science |
| 3 | Sonya | Material Science |
| 4 | Johnny | Music |
| 5 | Johnny | Statistics |
data_skill| Major | Skills | |
|---|---|---|
| 0 | Statistics | Inference |
| 1 | Computer Science | Machine learning |
| 2 | Material Science | Structure prediction |
| 3 | Music | Opera |
pd.merge(data_student,data_skill)| Student | Major | Skills | |
|---|---|---|---|
| 0 | Kitana | Statistics | Inference |
| 1 | Johnny | Statistics | Inference |
| 2 | Kitana | Computer Science | Machine learning |
| 3 | Jax | Computer Science | Machine learning |
| 4 | Sonya | Material Science | Structure prediction |
| 5 | Johnny | Music | Opera |
8.2.1.3 One-to-many join
Each row in one dataset is linked (or related) to one, or more rows in another dataset based on the key column(s).
data_student = pd.read_csv('./Datasets/student_one.csv')
data_skill = pd.read_csv('./Datasets/skills_many.csv')data_student| Student | Major | |
|---|---|---|
| 0 | Kitana | Statistics |
| 1 | Jax | Computer Science |
| 2 | Sonya | Material Science |
| 3 | Johnny | Music |
data_skill| Major | Skills | |
|---|---|---|
| 0 | Statistics | Inference |
| 1 | Statistics | Modeling |
| 2 | Computer Science | Machine learning |
| 3 | Computer Science | Computing |
| 4 | Material Science | Structure prediction |
| 5 | Music | Opera |
| 6 | Music | Pop |
| 7 | Music | Classical |
pd.merge(data_student,data_skill)| Student | Major | Skills | |
|---|---|---|---|
| 0 | Kitana | Statistics | Inference |
| 1 | Kitana | Statistics | Modeling |
| 2 | Jax | Computer Science | Machine learning |
| 3 | Jax | Computer Science | Computing |
| 4 | Sonya | Material Science | Structure prediction |
| 5 | Johnny | Music | Opera |
| 6 | Johnny | Music | Pop |
| 7 | Johnny | Music | Classical |
8.2.1.4 Many-to-many join
One, or more, rows in one dataset is linked (or related) to one, or more, rows in another dataset using the key column(s).
data_student = pd.read_csv('./Datasets/student_many.csv')
data_skill = pd.read_csv('./Datasets/skills_many.csv')data_student| Student | Major | |
|---|---|---|
| 0 | Kitana | Statistics |
| 1 | Kitana | Computer Science |
| 2 | Jax | Computer Science |
| 3 | Sonya | Material Science |
| 4 | Johnny | Music |
| 5 | Johnny | Statistics |
data_skill| Major | Skills | |
|---|---|---|
| 0 | Statistics | Inference |
| 1 | Statistics | Modeling |
| 2 | Computer Science | Machine learning |
| 3 | Computer Science | Computing |
| 4 | Material Science | Structure prediction |
| 5 | Music | Opera |
| 6 | Music | Pop |
| 7 | Music | Classical |
pd.merge(data_student,data_skill)| Student | Major | Skills | |
|---|---|---|---|
| 0 | Kitana | Statistics | Inference |
| 1 | Kitana | Statistics | Modeling |
| 2 | Johnny | Statistics | Inference |
| 3 | Johnny | Statistics | Modeling |
| 4 | Kitana | Computer Science | Machine learning |
| 5 | Kitana | Computer Science | Computing |
| 6 | Jax | Computer Science | Machine learning |
| 7 | Jax | Computer Science | Computing |
| 8 | Sonya | Material Science | Structure prediction |
| 9 | Johnny | Music | Opera |
| 10 | Johnny | Music | Pop |
| 11 | Johnny | Music | Classical |
Note that there are two ‘Statistics’ rows in data_student, and two ‘Statistics’ rows in data_skill, resulting in 2x2 = 4 ‘Statistics’ rows in the merged data. The same is true for the ‘Computer Science’ Major.
8.2.2 Join types with how argument
The above mentioned types of join (one-to-one, many-to-one, etc.) occur depening on the structure of the datasets being merged. We don’t have control over the type of join. However, we can control how the joins are occurring. We can merge (or join) two datasets in one of the following four ways:
innerjoin, (ii)leftjoin, (iii)rightjoin, (iv)outerjoin
8.2.2.1 inner join
This is the join that occurs by default, i.e., without specifying the how argument in the merge() function. In inner join, only those observations are merged that have the same value(s) in the key column(s) of both the datasets.
data_student = pd.read_csv('./Datasets/student_how.csv')
data_skill = pd.read_csv('./Datasets/skills_how.csv')data_student| Student | Major | |
|---|---|---|
| 0 | Kitana | Statistics |
| 1 | Jax | Computer Science |
| 2 | Sonya | Material Science |
data_skill| Major | Skills | |
|---|---|---|
| 0 | Statistics | Inference |
| 1 | Computer Science | Machine learning |
| 2 | Music | Opera |
pd.merge(data_student,data_skill)| Student | Major | Skills | |
|---|---|---|---|
| 0 | Kitana | Statistics | Inference |
| 1 | Jax | Computer Science | Machine learning |
When you may use inner join? You should use inner join when you cannot carry out the analysis unless the observation corresponding to the key column(s) is present in both the tables.
Example: Suppose you wish to analyze the association between vaccinations and covid infection rate based on country-level data. In one of the datasets, you have the infection rate for each country, while in the other one you have the number of vaccinations in each country. The countries which have either the vaccination or the infection rate missing, cannot help analyze the association. In such as case you may be interested only in countries that have values for both the variables. Thus, you will use inner join to discard the countries with either value missing.
8.2.2.2 left join
In left join, the merged dataset will have all the rows of the dataset that is specified first in the merge() function. Only those observations of the other dataset will be merged whose value(s) in the key column(s) exist in the dataset specified first in the merge() function.
pd.merge(data_student,data_skill,how='left')| Student | Major | Skills | |
|---|---|---|---|
| 0 | Kitana | Statistics | Inference |
| 1 | Jax | Computer Science | Machine learning |
| 2 | Sonya | Material Science | NaN |
When you may use left join? You should use left join when the primary variable(s) of interest are present in the one of the datasets, and whose missing values cannot be imputed. The variable(s) in the other dataset may not be as important or it may be possible to reasonably impute their values, if missing corresponding to the observation in the primary dataset.
Examples:
Suppose you wish to analyze the association between the covid infection rate and the government effectiveness score (a metric used to determine the effectiveness of the government in implementing policies, upholding law and order etc.) based on the data of all countries. Let us say that one of the datasets contains the covid infection rate, while the other one contains the government effectiveness score for each country. If the infection rate for a country is missing, it might be hard to impute. However, the government effectiveness score may be easier to impute based on GDP per capita, crime rate etc. - information that is easily available online. In such a case, you may wish to use a left join where you keep all the countries for which the infection rate is known.
Suppose you wish to analyze the association between demographics such as age, income etc. and the amount of credit card spend. Let us say one of the datasets contains the demographic information of each customer, while the other one contains the credit card spend for the customers who made at least one purchase. In such as case, you may want to do a left join as customers not making any purchase might be absent in the card spend data. Their spend can be imputed as zero after merging the datasets.
8.2.2.3 right join
In right join, the merged dataset will have all the rows of the dataset that is specified second in the merge() function. Only those observations of the other dataset will be merged whose value(s) in the key column(s) exist in the dataset specified second in the merge() function.
pd.merge(data_student,data_skill,how='right')| Student | Major | Skills | |
|---|---|---|---|
| 0 | Kitana | Statistics | Inference |
| 1 | Jax | Computer Science | Machine learning |
| 2 | NaN | Music | Opera |
When you may use right join? You can always use a left join instead of a right join. Their purpose is the same.
8.2.2.4 outer join
In outer join, the merged dataset will have all the rows of both the datasets being merged.
pd.merge(data_student,data_skill,how='outer')| Student | Major | Skills | |
|---|---|---|---|
| 0 | Kitana | Statistics | Inference |
| 1 | Jax | Computer Science | Machine learning |
| 2 | Sonya | Material Science | NaN |
| 3 | NaN | Music | Opera |
When you may use outer join? You should use an outer join when you cannot afford to lose data present in either of the tables. All the other joins may result in loss of data.
Example: Suppose I took two course surveys for this course. If I need to analyze student sentiment during the course, I will take an outer join of both the surveys. Assume that each survey is a dataset, where each row corresponds to a unique student. Even if a student has answered one of the two surverys, it will be indicative of the sentiment, and will be useful to keep in the merged dataset.
8.3 Concatenating datasets
The Pandas DataFrame method concat() is used to stack datasets along an axis. The method is similar to NumPy’s concatenate() method.
Example: You are given the life expectancy data of each continent as a separate *.csv file. Visualize the change of life expectancy over time for different continents.
data_asia = pd.read_csv('./Datasets/gdp_lifeExpec_Asia.csv')
data_europe = pd.read_csv('./Datasets/gdp_lifeExpec_Europe.csv')
data_africa = pd.read_csv('./Datasets/gdp_lifeExpec_Africa.csv')
data_oceania = pd.read_csv('./Datasets/gdp_lifeExpec_Oceania.csv')
data_americas = pd.read_csv('./Datasets/gdp_lifeExpec_Americas.csv')#Appending all the data files, i.e., stacking them on top of each other
data_all_continents = pd.concat([data_asia,data_europe,data_africa,data_oceania,data_americas],keys = ['Asia','Europe','Africa','Oceania','Americas'])
data_all_continents| country | year | lifeExp | pop | gdpPercap | ||
|---|---|---|---|---|---|---|
| Asia | 0 | Afghanistan | 1952 | 28.801 | 8425333 | 779.445314 |
| 1 | Afghanistan | 1957 | 30.332 | 9240934 | 820.853030 | |
| 2 | Afghanistan | 1962 | 31.997 | 10267083 | 853.100710 | |
| 3 | Afghanistan | 1967 | 34.020 | 11537966 | 836.197138 | |
| 4 | Afghanistan | 1972 | 36.088 | 13079460 | 739.981106 | |
| ... | ... | ... | ... | ... | ... | ... |
| Americas | 295 | Venezuela | 1987 | 70.190 | 17910182 | 9883.584648 |
| 296 | Venezuela | 1992 | 71.150 | 20265563 | 10733.926310 | |
| 297 | Venezuela | 1997 | 72.146 | 22374398 | 10165.495180 | |
| 298 | Venezuela | 2002 | 72.766 | 24287670 | 8605.047831 | |
| 299 | Venezuela | 2007 | 73.747 | 26084662 | 11415.805690 |
1704 rows × 5 columns
Let’s have the continent as a column as we need to use that in the visualization.
data_all_continents.reset_index(inplace = True)data_all_continents.head()| level_0 | level_1 | country | year | lifeExp | pop | gdpPercap | |
|---|---|---|---|---|---|---|---|
| 0 | Asia | 0 | Afghanistan | 1952 | 28.801 | 8425333 | 779.445314 |
| 1 | Asia | 1 | Afghanistan | 1957 | 30.332 | 9240934 | 820.853030 |
| 2 | Asia | 2 | Afghanistan | 1962 | 31.997 | 10267083 | 853.100710 |
| 3 | Asia | 3 | Afghanistan | 1967 | 34.020 | 11537966 | 836.197138 |
| 4 | Asia | 4 | Afghanistan | 1972 | 36.088 | 13079460 | 739.981106 |
data_all_continents.drop(columns = 'level_1',inplace = True)
data_all_continents.rename(columns = {'level_0':'continent'},inplace = True)
data_all_continents.head()| continent | country | year | lifeExp | pop | gdpPercap | |
|---|---|---|---|---|---|---|
| 0 | Asia | Afghanistan | 1952 | 28.801 | 8425333 | 779.445314 |
| 1 | Asia | Afghanistan | 1957 | 30.332 | 9240934 | 820.853030 |
| 2 | Asia | Afghanistan | 1962 | 31.997 | 10267083 | 853.100710 |
| 3 | Asia | Afghanistan | 1967 | 34.020 | 11537966 | 836.197138 |
| 4 | Asia | Afghanistan | 1972 | 36.088 | 13079460 | 739.981106 |
#change of life expectancy over time for different continents
a = sns.FacetGrid(data_all_continents,col = 'continent',col_wrap = 3,height = 4.5,aspect = 1)#height = 3,aspect = 0.8)
a.map(sns.lineplot,'year','lifeExp')
a.add_legend()
In the above example, datasets were appended (or stacked on top of each other).
Datasets can also be concatenated side-by-side (by providing the argument axis = 1 with the concat() function) as we saw with the merge function.
8.3.1 Practice exercise 4
Read the documentations of the Pandas DataFrame methods merge() and concat(), and identify the differences. Mention examples when you can use (i) either, (ii) only concat(), (iii) only merge()
Solution:
If we need to merge datasets using row indices, we can use either function.
If we need to stack datasets one on top of the other, we can only use
concat()If we need to merge datasets using overlapping columns we can only use
merge()
8.4 Reshaping data
Data often needs to be re-arranged to ease analysis.
8.4.1 Pivoting “long” to “wide” format
pivot()
This function helps re-arrange data from the ‘long’ form to a ‘wide’ form.
Example: Let us consider the dataset data_all_continents obtained in the previous section after concatenating the data of all the continents.
data_all_continents.head()| continent | country | year | lifeExp | pop | gdpPercap | |
|---|---|---|---|---|---|---|
| 0 | Asia | Afghanistan | 1952 | 28.801 | 8425333 | 779.445314 |
| 1 | Asia | Afghanistan | 1957 | 30.332 | 9240934 | 820.853030 |
| 2 | Asia | Afghanistan | 1962 | 31.997 | 10267083 | 853.100710 |
| 3 | Asia | Afghanistan | 1967 | 34.020 | 11537966 | 836.197138 |
| 4 | Asia | Afghanistan | 1972 | 36.088 | 13079460 | 739.981106 |
8.4.1.1 Pivoting a single column
For visualizing life expectancy in 2007 against life expectancy in 1957, we will need to filter the data, and then make the plot. Everytime that we need to compare a metric for a year against another year, we will need to filter the data.
If we need to often compare metrics of a year against another year, it will be easier to have each year as a separate column, instead of having all years in a single column.
As we are increasing the number of columns and decreasing the number of rows, we are re-arranging the data from long-form to wide-form.
data_wide = data_all_continents.pivot(index = ['continent','country'],columns = 'year',values = 'lifeExp')data_wide.head()| year | 1952 | 1957 | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | 2002 | 2007 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| continent | country | ||||||||||||
| Africa | Algeria | 43.077 | 45.685 | 48.303 | 51.407 | 54.518 | 58.014 | 61.368 | 65.799 | 67.744 | 69.152 | 70.994 | 72.301 |
| Angola | 30.015 | 31.999 | 34.000 | 35.985 | 37.928 | 39.483 | 39.942 | 39.906 | 40.647 | 40.963 | 41.003 | 42.731 | |
| Benin | 38.223 | 40.358 | 42.618 | 44.885 | 47.014 | 49.190 | 50.904 | 52.337 | 53.919 | 54.777 | 54.406 | 56.728 | |
| Botswana | 47.622 | 49.618 | 51.520 | 53.298 | 56.024 | 59.319 | 61.484 | 63.622 | 62.745 | 52.556 | 46.634 | 50.728 | |
| Burkina Faso | 31.975 | 34.906 | 37.814 | 40.697 | 43.591 | 46.137 | 48.122 | 49.557 | 50.260 | 50.324 | 50.650 | 52.295 |
With values of year as columns, it is easy to compare any metric for different years.
#visualizing the change in life expectancy of all countries in 2007 as compared to that in 1957, i.e., the overall change in life expectancy in 50 years.
sns.scatterplot(data = data_wide, x = 1957,y=2007,hue = 'continent')
sns.lineplot(data = data_wide, x = 1957,y = 1957)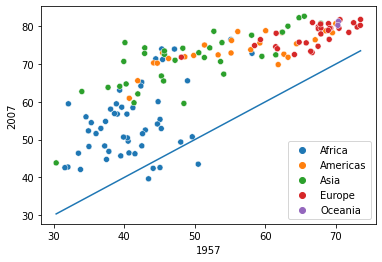
Observe that for some African countries, the life expectancy has decreased after 50 years. It is worth investigating these countries to identify factors associated with the decrease.
8.4.1.2 Pivoting multiple columns
In the above transformation, we retained only lifeExp in the ‘wide’ dataset. Suppose, we are also interested in visualizing GDP per capita of countries in one year against another year. In that case, we must have gdpPercap in the ’wide’-form data as well.
Let us create a dataset named as data_wide_lifeExp_gdpPercap that will contain both lifeExp and gdpPercap for each year in a separate column. We will specify the columns to pivot in the values argument of the pivot() function.
data_wide_lifeExp_gdpPercap = data_all_continents.pivot(index = ['continent','country'],columns = 'year',values = ['lifeExp','gdpPercap'])
data_wide_lifeExp_gdpPercap.head()| lifeExp | ... | gdpPercap | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| year | 1952 | 1957 | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | ... | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | 2002 | 2007 | |
| continent | country | |||||||||||||||||||||
| Africa | Algeria | 43.077 | 45.685 | 48.303 | 51.407 | 54.518 | 58.014 | 61.368 | 65.799 | 67.744 | 69.152 | ... | 2550.816880 | 3246.991771 | 4182.663766 | 4910.416756 | 5745.160213 | 5681.358539 | 5023.216647 | 4797.295051 | 5288.040382 | 6223.367465 |
| Angola | 30.015 | 31.999 | 34.000 | 35.985 | 37.928 | 39.483 | 39.942 | 39.906 | 40.647 | 40.963 | ... | 4269.276742 | 5522.776375 | 5473.288005 | 3008.647355 | 2756.953672 | 2430.208311 | 2627.845685 | 2277.140884 | 2773.287312 | 4797.231267 | |
| Benin | 38.223 | 40.358 | 42.618 | 44.885 | 47.014 | 49.190 | 50.904 | 52.337 | 53.919 | 54.777 | ... | 949.499064 | 1035.831411 | 1085.796879 | 1029.161251 | 1277.897616 | 1225.856010 | 1191.207681 | 1232.975292 | 1372.877931 | 1441.284873 | |
| Botswana | 47.622 | 49.618 | 51.520 | 53.298 | 56.024 | 59.319 | 61.484 | 63.622 | 62.745 | 52.556 | ... | 983.653976 | 1214.709294 | 2263.611114 | 3214.857818 | 4551.142150 | 6205.883850 | 7954.111645 | 8647.142313 | 11003.605080 | 12569.851770 | |
| Burkina Faso | 31.975 | 34.906 | 37.814 | 40.697 | 43.591 | 46.137 | 48.122 | 49.557 | 50.260 | 50.324 | ... | 722.512021 | 794.826560 | 854.735976 | 743.387037 | 807.198586 | 912.063142 | 931.752773 | 946.294962 | 1037.645221 | 1217.032994 | |
5 rows × 24 columns
The metric for each year is now in a separate column, and can be visualized directly. Note that re-arranging the dataset from the ‘long’-form to ‘wide-form’ leads to hierarchical indexing of columns when multiple ‘values’ need to be re-arranged. In this case, the multiple ‘values’ that need to be re-arranged are lifeExp and gdpPercap.
8.4.2 Melting “wide” to “long” format
melt()
This function is used to re-arrange the dataset from the ‘wide’ form to the ‘long’ form.
8.4.2.1 Melting columns with a single type of value
Let us consider data_wide created in the previous section.
data_wide.head()| year | 1952 | 1957 | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | 2002 | 2007 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| continent | country | ||||||||||||
| Africa | Algeria | 43.077 | 45.685 | 48.303 | 51.407 | 54.518 | 58.014 | 61.368 | 65.799 | 67.744 | 69.152 | 70.994 | 72.301 |
| Angola | 30.015 | 31.999 | 34.000 | 35.985 | 37.928 | 39.483 | 39.942 | 39.906 | 40.647 | 40.963 | 41.003 | 42.731 | |
| Benin | 38.223 | 40.358 | 42.618 | 44.885 | 47.014 | 49.190 | 50.904 | 52.337 | 53.919 | 54.777 | 54.406 | 56.728 | |
| Botswana | 47.622 | 49.618 | 51.520 | 53.298 | 56.024 | 59.319 | 61.484 | 63.622 | 62.745 | 52.556 | 46.634 | 50.728 | |
| Burkina Faso | 31.975 | 34.906 | 37.814 | 40.697 | 43.591 | 46.137 | 48.122 | 49.557 | 50.260 | 50.324 | 50.650 | 52.295 |
Suppose, we wish to visualize the change of life expectancy over time for different continents, as we did in section 8.3. For plotting lifeExp against year, all the years must be in a single column. Thus, we need to melt the columns of data_wide to a single column and call it year.
But before melting the columns in the above dataset, we will convert continent to a column, as we need to make subplots based on continent.
The Pandas DataFrame method reset_index() can be used to remove one or more levels of indexing from the DataFrame.
#Making 'continent' a column instead of row-index at level 0
data_wide.reset_index(inplace=True,level=0)
data_wide.head()| year | continent | 1952 | 1957 | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | 2002 | 2007 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| country | |||||||||||||
| Algeria | Africa | 43.077 | 45.685 | 48.303 | 51.407 | 54.518 | 58.014 | 61.368 | 65.799 | 67.744 | 69.152 | 70.994 | 72.301 |
| Angola | Africa | 30.015 | 31.999 | 34.000 | 35.985 | 37.928 | 39.483 | 39.942 | 39.906 | 40.647 | 40.963 | 41.003 | 42.731 |
| Benin | Africa | 38.223 | 40.358 | 42.618 | 44.885 | 47.014 | 49.190 | 50.904 | 52.337 | 53.919 | 54.777 | 54.406 | 56.728 |
| Botswana | Africa | 47.622 | 49.618 | 51.520 | 53.298 | 56.024 | 59.319 | 61.484 | 63.622 | 62.745 | 52.556 | 46.634 | 50.728 |
| Burkina Faso | Africa | 31.975 | 34.906 | 37.814 | 40.697 | 43.591 | 46.137 | 48.122 | 49.557 | 50.260 | 50.324 | 50.650 | 52.295 |
data_melted=pd.melt(data_wide,id_vars = ['continent'],var_name = 'Year',value_name = 'LifeExp')
data_melted.head()| continent | Year | LifeExp | |
|---|---|---|---|
| 0 | Africa | 1952 | 43.077 |
| 1 | Africa | 1952 | 30.015 |
| 2 | Africa | 1952 | 38.223 |
| 3 | Africa | 1952 | 47.622 |
| 4 | Africa | 1952 | 31.975 |
With the above DataFrame, we can visualize the mean life expectancy against year separately for each continent.
If we wish to have country also in the above data, we can keep it while resetting the index:
#Creating 'data_wide' again
data_wide = data_all_continents.pivot(index = ['continent','country'],columns = 'year',values = 'lifeExp')
#Resetting the row-indices to default values
data_wide.reset_index(inplace=True)
data_wide.head()| year | continent | country | 1952 | 1957 | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | 2002 | 2007 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Africa | Algeria | 43.077 | 45.685 | 48.303 | 51.407 | 54.518 | 58.014 | 61.368 | 65.799 | 67.744 | 69.152 | 70.994 | 72.301 |
| 1 | Africa | Angola | 30.015 | 31.999 | 34.000 | 35.985 | 37.928 | 39.483 | 39.942 | 39.906 | 40.647 | 40.963 | 41.003 | 42.731 |
| 2 | Africa | Benin | 38.223 | 40.358 | 42.618 | 44.885 | 47.014 | 49.190 | 50.904 | 52.337 | 53.919 | 54.777 | 54.406 | 56.728 |
| 3 | Africa | Botswana | 47.622 | 49.618 | 51.520 | 53.298 | 56.024 | 59.319 | 61.484 | 63.622 | 62.745 | 52.556 | 46.634 | 50.728 |
| 4 | Africa | Burkina Faso | 31.975 | 34.906 | 37.814 | 40.697 | 43.591 | 46.137 | 48.122 | 49.557 | 50.260 | 50.324 | 50.650 | 52.295 |
#Melting the 'year' column
data_melted=pd.melt(data_wide,id_vars = ['continent','country'],var_name = 'Year',value_name = 'LifeExp')
data_melted.head()| continent | country | Year | LifeExp | |
|---|---|---|---|---|
| 0 | Africa | Algeria | 1952 | 43.077 |
| 1 | Africa | Angola | 1952 | 30.015 |
| 2 | Africa | Benin | 1952 | 38.223 |
| 3 | Africa | Botswana | 1952 | 47.622 |
| 4 | Africa | Burkina Faso | 1952 | 31.975 |
8.4.2.2 Melting columns with multiple types of values
Consider the dataset created in Section 8.4.1.2. It has two types of values - lifeExp and gdpPercapita, which are the column labels at the outer level. The melt() function will melt all the years of data into a single column. However, it will create another column based on the outer level column labels - lifeExp and gdpPercapita to distinguish between these two types of values. Here, we see that the function melt() internally uses hierarchical indexing to handle the transformation of multiple types of columns.
data_melt = pd.melt(data_wide_lifeExp_gdpPercap.reset_index(),id_vars = ['continent','country'],var_name = ['Metric','year'])
data_melt.head()| continent | country | Metric | year | value | |
|---|---|---|---|---|---|
| 0 | Africa | Algeria | lifeExp | 1952 | 43.077 |
| 1 | Africa | Angola | lifeExp | 1952 | 30.015 |
| 2 | Africa | Benin | lifeExp | 1952 | 38.223 |
| 3 | Africa | Botswana | lifeExp | 1952 | 47.622 |
| 4 | Africa | Burkina Faso | lifeExp | 1952 | 31.975 |
Although the data above is in ‘long’-form, it is not quiet in its original format, as in data_all_continents. We need to pivot again by Metric to have two separate columns of gdpPercap and lifeExp.
data_restore = data_melt.pivot(index = ['continent','country','year'],columns = 'Metric')
data_restore.head()| value | ||||
|---|---|---|---|---|
| Metric | gdpPercap | lifeExp | ||
| continent | country | year | ||
| Africa | Algeria | 1952 | 2449.008185 | 43.077 |
| 1957 | 3013.976023 | 45.685 | ||
| 1962 | 2550.816880 | 48.303 | ||
| 1967 | 3246.991771 | 51.407 | ||
| 1972 | 4182.663766 | 54.518 | ||
Now, we can convert the row indices of continent and country to columns to restore the dataset to the same form as data_all_continents.
data_restore.reset_index(inplace = True)
data_restore.head()| continent | country | year | value | ||
|---|---|---|---|---|---|
| Metric | gdpPercap | lifeExp | |||
| 0 | Africa | Algeria | 1952 | 2449.008185 | 43.077 |
| 1 | Africa | Algeria | 1957 | 3013.976023 | 45.685 |
| 2 | Africa | Algeria | 1962 | 2550.816880 | 48.303 |
| 3 | Africa | Algeria | 1967 | 3246.991771 | 51.407 |
| 4 | Africa | Algeria | 1972 | 4182.663766 | 54.518 |
8.4.3 Practice exercise 5
8.4.3.1
Both unstack() and pivot() seem to transform the data from the ‘long’ form to the ‘wide’ form. Is there a difference between the two functions?
Solution:
Yes, both the functions transform the data from the ‘long’ form to the ‘wide’ form. However, unstack() pivots the row indices, while pivot() pivots the columns of the DataFrame.
Even though both functions are a bit different, it is possible to just use one of them to perform a reshaping operation. If we wish to pivot a column, we can either use pivot() directly on the column, or we can convert the column to row indices and then use unstack(). If we wish to pivot row indices, we can either use unstack() directly on the row indices, or we can convert row indices to a column and then use pivot().
To summarise, using one function may be more straightforward than using the other one, but either can be used for reshaping data from the ‘long’ form to the ‘wide’ form.
Below is an example where we perform the same reshaping operation with either function.
Consider the data data_all_continent. Suppose we wish to transform it to data_wide as we did using pivot() in Section 8.4.1.1. Let us do it using unstack(), instead of pivot().
The first step will be to reindex data to set year as row indices, and also continent and country as row indices because these two column were set as indices with the pivot() function in Section 8.4.1.1.
#Reindexing data to make 'continent', 'country', and 'year' as hierarchical row indices
data_reindexed=data_all_continents.set_index(['continent','country','year'])
data_reindexed| lifeExp | pop | gdpPercap | |||
|---|---|---|---|---|---|
| continent | country | year | |||
| Asia | Afghanistan | 1952 | 28.801 | 8425333 | 779.445314 |
| 1957 | 30.332 | 9240934 | 820.853030 | ||
| 1962 | 31.997 | 10267083 | 853.100710 | ||
| 1967 | 34.020 | 11537966 | 836.197138 | ||
| 1972 | 36.088 | 13079460 | 739.981106 | ||
| ... | ... | ... | ... | ... | ... |
| Americas | Venezuela | 1987 | 70.190 | 17910182 | 9883.584648 |
| 1992 | 71.150 | 20265563 | 10733.926310 | ||
| 1997 | 72.146 | 22374398 | 10165.495180 | ||
| 2002 | 72.766 | 24287670 | 8605.047831 | ||
| 2007 | 73.747 | 26084662 | 11415.805690 |
1704 rows × 3 columns
Now we can use unstack() to pivot the desired row index, i.e., year. Also, since we are only interested in pivoting the values of lifeExp (as in the example in Section 8.4.1.1), we will filter the pivoted data with the lifeExp column label.
data_wide_with_unstack=data_reindexed.unstack('year')['lifeExp']
data_wide_with_unstack| year | 1952 | 1957 | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | 2002 | 2007 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| continent | country | ||||||||||||
| Africa | Algeria | 43.077 | 45.685 | 48.303 | 51.407 | 54.518 | 58.014 | 61.368 | 65.799 | 67.744 | 69.152 | 70.994 | 72.301 |
| Angola | 30.015 | 31.999 | 34.000 | 35.985 | 37.928 | 39.483 | 39.942 | 39.906 | 40.647 | 40.963 | 41.003 | 42.731 | |
| Benin | 38.223 | 40.358 | 42.618 | 44.885 | 47.014 | 49.190 | 50.904 | 52.337 | 53.919 | 54.777 | 54.406 | 56.728 | |
| Botswana | 47.622 | 49.618 | 51.520 | 53.298 | 56.024 | 59.319 | 61.484 | 63.622 | 62.745 | 52.556 | 46.634 | 50.728 | |
| Burkina Faso | 31.975 | 34.906 | 37.814 | 40.697 | 43.591 | 46.137 | 48.122 | 49.557 | 50.260 | 50.324 | 50.650 | 52.295 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Europe | Switzerland | 69.620 | 70.560 | 71.320 | 72.770 | 73.780 | 75.390 | 76.210 | 77.410 | 78.030 | 79.370 | 80.620 | 81.701 |
| Turkey | 43.585 | 48.079 | 52.098 | 54.336 | 57.005 | 59.507 | 61.036 | 63.108 | 66.146 | 68.835 | 70.845 | 71.777 | |
| United Kingdom | 69.180 | 70.420 | 70.760 | 71.360 | 72.010 | 72.760 | 74.040 | 75.007 | 76.420 | 77.218 | 78.471 | 79.425 | |
| Oceania | Australia | 69.120 | 70.330 | 70.930 | 71.100 | 71.930 | 73.490 | 74.740 | 76.320 | 77.560 | 78.830 | 80.370 | 81.235 |
| New Zealand | 69.390 | 70.260 | 71.240 | 71.520 | 71.890 | 72.220 | 73.840 | 74.320 | 76.330 | 77.550 | 79.110 | 80.204 |
142 rows × 12 columns
The above dataset is the same as that obtained using the pivot() function in Section 8.4.1.1.
8.4.3.2
Both stack() and melt() seem to transform the data from the ‘wide’ form to the ‘long’ form. Is there a difference between the two functions?
Solution:
Following the trend of the previous question, we can always use stack() instead of melt() and vice-versa. The main difference is that melt() lets us choose the indentifier columns with the argument id_vars. However, if we use stack(), we will need to set the relevant melted row indices as columns. On the other hand, if we wished to have the melted columns as row indices, we can either directly use stack() or use melt() and then set the desired columns as row indices.
To summarise, using one function may be more straightforward than using the other one, but either can be used for reshaping data from the ‘wide’ form to the ‘long’ form.
Let us melt the data data_wide_with_unstack using the stack() function to obtain the same dataset as obtained with the melt() function in Section 8.4.1.2.
#Stacking the data
data_stacked = data_wide_with_unstack.stack()
data_stackedcontinent country year
Africa Algeria 1952 43.077
1957 45.685
1962 48.303
1967 51.407
1972 54.518
...
Oceania New Zealand 1987 74.320
1992 76.330
1997 77.550
2002 79.110
2007 80.204
Length: 1704, dtype: float64Now we need to convert the row indices continent and country to columns as in the melted data in Section 8.4.1.2.
#Putting 'continent' and 'country' as columns
data_long_with_stack = data_stacked.reset_index()
data_long_with_stack| continent | country | year | 0 | |
|---|---|---|---|---|
| 0 | Africa | Algeria | 1952 | 43.077 |
| 1 | Africa | Algeria | 1957 | 45.685 |
| 2 | Africa | Algeria | 1962 | 48.303 |
| 3 | Africa | Algeria | 1967 | 51.407 |
| 4 | Africa | Algeria | 1972 | 54.518 |
| ... | ... | ... | ... | ... |
| 1699 | Oceania | New Zealand | 1987 | 74.320 |
| 1700 | Oceania | New Zealand | 1992 | 76.330 |
| 1701 | Oceania | New Zealand | 1997 | 77.550 |
| 1702 | Oceania | New Zealand | 2002 | 79.110 |
| 1703 | Oceania | New Zealand | 2007 | 80.204 |
1704 rows × 4 columns
Finally, we need to rename the column named as 0 to LifeExp to obtain the same dataset as in Section 8.4.1.2.
#Renaming column 0 to 'LifeExp'
data_long_with_stack.rename(columns = {0:'LifeExp'},inplace=True)
data_long_with_stack| continent | country | year | LifeExp | |
|---|---|---|---|---|
| 0 | Africa | Algeria | 1952 | 43.077 |
| 1 | Africa | Algeria | 1957 | 45.685 |
| 2 | Africa | Algeria | 1962 | 48.303 |
| 3 | Africa | Algeria | 1967 | 51.407 |
| 4 | Africa | Algeria | 1972 | 54.518 |
| ... | ... | ... | ... | ... |
| 1699 | Oceania | New Zealand | 1987 | 74.320 |
| 1700 | Oceania | New Zealand | 1992 | 76.330 |
| 1701 | Oceania | New Zealand | 1997 | 77.550 |
| 1702 | Oceania | New Zealand | 2002 | 79.110 |
| 1703 | Oceania | New Zealand | 2007 | 80.204 |
1704 rows × 4 columns
